Медиатека
Quantitative Assay of SARS-CoV-2 RNA and Level of Proinflammatory Protein Gene Transcripts in Peripheral Blood Leukocytes after a Novel Coronavirus Infection
Статья «Количественный анализ РНК SARS-CoV-2 и уровня транскриптов генов провоспалительных белков в периферических лейкоцитах крови после новой коронавирусной инфекции».
Autors: L. V. Topchieva 1, 2, O. V. Balan 1, 2, A. V. Men’shenin 3, I. E. Malysheva 1, 2, and E. L. Tikhonovich 2
Translated from Byulleten’ Eksperimental’noi Biologii i Meditsiny, Vol. 173, No. 6, pp. 726-731, June, 2022 Original article submitted April 20, 2022
The possibility of finding persistent SARS-CoV-2 viral particles in human peripheral blood leukocytes after a novel coronavirus infection was shown. The results of droplet digital PCR showed that 19 of 24 examined subjects had from 4 to 555 copies of the Nsp4 SARS-CoV-2 gene in 5-6 months after infection. The presence of this transcript in peripheral blood leukocytes was associated with reduced expression of FOXP3 gene and increased level of RORγ gene mRNA. The copy number of the Nsp4 gene negatively correlated with the level of FOXP3 gene mRNA (r=-0.45; p=0.028), but showed a positive correlation with the DANCR long non-coding RNA (r=0.94; p<0.001). In SARS-CoV-2-positive healthy individuals, the level of TLR2, NLRP3, and IL1B gene transcripts was higher than in SARS-CoV-2-negative donors. The presence of SARS-CoV-2 in a persistent form is probably associated with impaired immu- nosuppression and the development of chronic inflammation in apparently healthy volunteers after a new coronavirus infection.
Key Words: SARS-CoV-2; novel coronavirus infection; droplet digital PCR; gene expression; inflammation
A number of viral infections are characterized by al- ternating latent and lytic (active) phases. For example, gamma-herpesviruses (Epstein—Barr virus, Saimiri her- pesvirus in monkeys, and HHV-8 herpes associated with Kaposi’s sarcoma in humans) demonstrate alternative life cycles (latent and lytic) upon infection 1. In the case of a latent or asymptomatic infection, such as herpes, influenza, after a primary infection or between relapses of the disease, the virus is hardly detected or not detected at all in biological fluids. Viruses can be reactivated in host cells when stimulated by various exogenous and/or endogenous factors, and this process is associated with changes in the transcriptional activity of the viral genome or the host genome. Thus, the transition of the Kaposi sarcoma virus HHV-8 from the latent to the lytic phase is the result of transcrip- tional changes in its genome caused by the replication and transcription activator (RTA) 1.
Since 2019, the main attention of infectious dis- ease specialists has been turned to the study of the emergence and spread of SARS-CoV-2, its epidemio- logy, diagnosis, clinical course, and treatment of the novel coronavirus disease COVID-19 caused by this virus. COVID-19 is a group of acute infectious dis- eases characterized by a syndrome of respiratory tract injury, in some cases occurring in a severe form with high mortality, and in some cases asymptomatic 2. Whether SARS-CoV-2 persists in immune or other body cells after patients with novel coronavirus in- fection recover remains unknown. There is evidence of long-term persistence of SARS-CoV-2 RNA (up to 4 months from the onset of the disease) in cells and blood sera, which may be due to the persistence of the pathogen or integration of SARS-CoV-2 DNA into human genome 3.
Virus persistence can affect the host’s immune sys- tem and can be accompanied by changes in the profile of inflammatory markers. For instance, in male Norwe- gian rats, the persistent phase of Hantavirus infection (Seoul virus, SEOV) in tissues supporting increased viral replication (i.e., in the lungs) was associated with reduced production of antiviral (IFNβ, IFNγ) and proinflammatory (IL-1β, IL-6, and TNFα) proteins 4. It has been shown that hantavirus persistence in ro- dents is associated with increased activity of regula- tory T cells 4. COVID-19 is often accompanied by lymphopenia with a significant decrease in the level of CD8 (T-cell differentiation marker) and an increase in the CD4/CD8 ratio, which allows predicting the severity of the disease 5. This leads to an immuno- suppressive state and can cause reactivation of other latent viral infections 6. It is unknown whether the presence of viral particles in the human body after a COVID-19 affects immune parameters and the content of inflammatory markers.
In the case of a latent phase of COVID-19, quan- tification of residual viruses in clinical specimens is important. Droplet digital PCR allows detecting targets with a concentration of 0.0001% copies, which cannot be detected by other methods.
The aim of the study is to quantify SARS-CoV-2 in peripheral blood leukocytes (PBL) from people re- covered from COVID-19 using quantitative real-time PCR (qPCR) and droplet digital PCR (ddPCR) and as- sess the level of transcripts of genes encoding T-cell differentiation markers and proinflammatory proteins.
1Institute of Biology, Karelian Research Centre, Russian Academy of Sciences, Petrozavodsk, Republic of Karelia, Russia; 2Center for Biomedical Research, Karelian Research Centre, Russian Academy of Sciences, Petrozavodsk, Republic of Karelia, Russia; 3North-West Subdivision of Helicon Company, LLC, St. Peters- burg, Russia. Address for correspondence: topchieva67@mail. ru. L. V. Topchieva
MATERIALS AND METHODS
Peripheral blood samples for the study were collected from healthy subjects without history of COVID-19 (n=28, mean age 52.0±2.4 years) and subjects reco- vered from COVID-19 (at least 5 months after recov- ery; n=24, mean age 50.0±2.7 years). In the latter group, 7 subjects had concomitant diseases: type 2 diabetes mellitus (3 cases) and chronic pancreatitis (4 cases). The material for the study was obtained with the assistance of the V. A. Baranov Republican Hospi- tal (Petrozavodsk, Russia) and Center for Biomedical Research, Karelian Research Centre. Written informed consent to conduct the research study was obtained from all participants (n=52, mean age 51.0±1.8 years). The study was approved by the Ethics Committee of the V. A. Baranov Republican Hospital and was carried out in compliance with the principles of the WMA Declaration of Helsinki.
Among those who were diagnosed with COVID-19 at the time of the disease, the disease manifested as an acute respiratory infection without lung damage in 18 subjects, was asymptomatic in 4 subjects, and was accompanied by lung damage with the development of community-acquired bilateral pneumonia in 2 patients. All participants were negative for SARS-CoV-2 at the time of the study.
To exclude the influence of inflammation, the ex- pression of genes of proinflammatory proteins and immune cell differentiation markers was assayed in PBL of apparently healthy volunteers (n=30, mean age 44.46±3.25 years), who according to the results of our study had a negative (14 people) or positive (16 people) the status of SARS-CoV-2 from the sample for detection of viral RNA by ddPCR. The exclusion criteria were the presence of concomitant immune-in- flammatory diseases, infectious diseases during the previous month, tobacco smoking, alcohol abuse, and body mass index ≥28 kg/m2.
Total RNA was isolated using PureZole reagent (Bio-Rad) from PBL obtained after treatment of whole blood with 0.86% ammonium chloride solution and centrifugation at 1250 rpm using Liston C 2201 cen- trifuge. The quality of total RNA was assessed by electrophoresis in 1% agarose gel. The content of total RNA was assessed on a SmartSpecPlus spectropho- tometer (Bio-Rad). Synthesis of complementary DNA was synthesized using the MMLV RT kit (Eurogen) on an RNA matrix pretreated with DNase (Synthol). qPCR was performed on a LightCycler amplifier (Roche). PCR mixture (25 µl) contained 5 µl qPCRmix-HS SYBR PCR mix (Eurogen), 10 pmol forward and reverse prim- ers, 30 ng complementary DNA, and 18 µl deionized sterile water. The 18S rRNA was used as a reference gene7. The specificity of PCR products was checked by melting curves. Each PCR was performed at least 3 times. The relative level of gene expression was assessed by ΔСt.
ddPCR was performed using Droplet Digital PCR QX200 system (Bio-Rad) and QX200 EvaGreen ddPCR supermix. Primers for PCR were designed using Bea- con Designer 5.0 software. The sequence of primers for assessing the level of transcripts of the FOXP3 and RORγ genes was presented elsewhere 8. The nucleo- tide sequence of other primers is shown in Table 1.
Statistical data processing was performed using the Statgraphics Centurion XVI 16.1.11 software pack- age. As according to the Shapiro—Wilk test, biochem- ical parameters did not fit normal distribution. Signif- icance of differences in the level of gene expression was assessed using the Mann—Whitney U test. The results are presented as Me (Q1; Q3). Analysis of vari- ance was carried out using the Kruskal—Wallis H test. The age of the individuals included in the study is presented as M±m. The differences were considered significant at p<0.05.
TABLE 1. Primer Sequence for qPCR
Note. The source of all presented primers is own design.
RESULTS
qPCR detected no copies of the gene sequence en- coding the SARS-CoV-2 Nsp4 protein in PBL of the examined subjects, while ddPCR detected copies of this sequence in PBL of 19 donors who had recovered from COVID-19. The number of copies per 1 µl ranged from 4 to 555. Five donors from the group without a history of COVID-19 (SARS-Cov-2 IgG antibodies were not tested) were positive and had 3 to 192 copies/µl. These results indicate the presence of SARS-CoV-2 virus in a latent form in PBL of individuals who have previously had this infection.
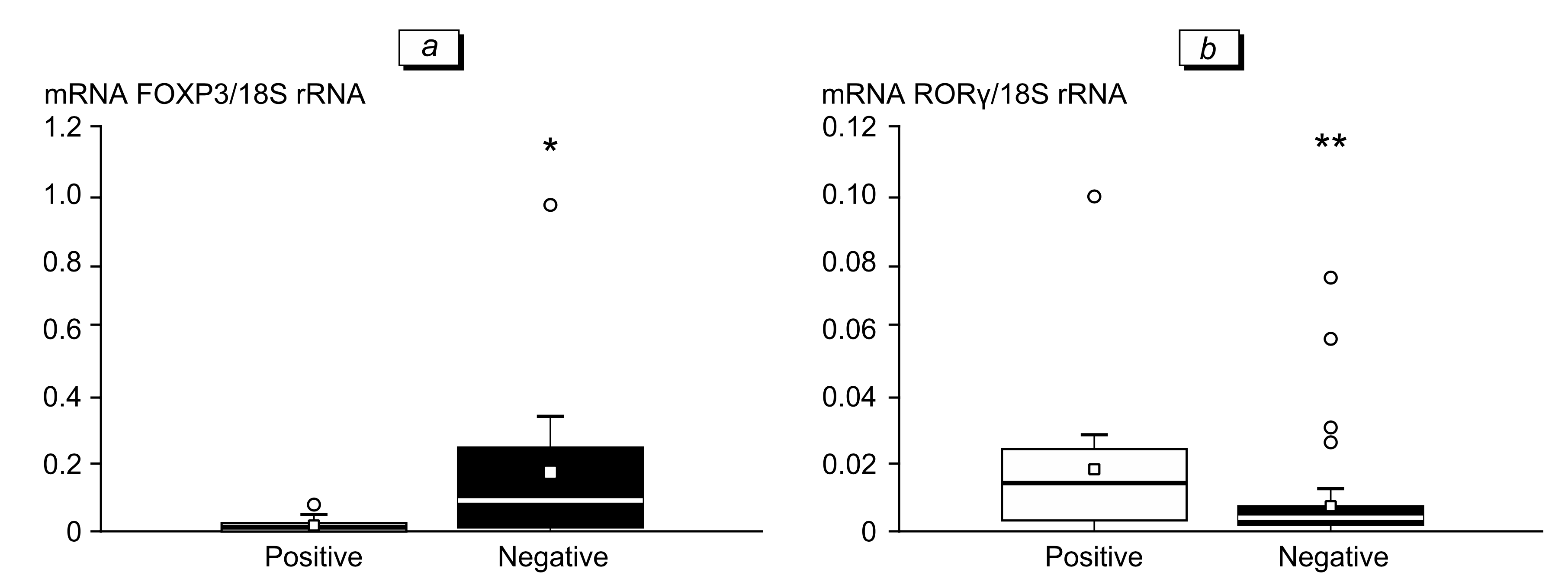
The presence of SARS-CoV-2 Nsp4 copies in PBL was accompanied by a decrease in transcriptional ac- tivity of the FOXP3 gene and an increase in the ex- pression of the RORγ gene (Fig. 1). The expression of FOXP3 and alpha subunit of the IL-2 receptor genes (IL2R) is the most important characteristic of T-reg- ulatory lymphocytes (Treg), while the gene encoding transcription factor RORγ is the most important mark- er of T-helpers 17 (Th17). Most likely that the pres- ence of virus copies affected the immune cells profile towards a decrease in the content of suppressor Treg cells and an increase in the number of proinflam- matory Th17. Similar changes in the CD4/Treg and Th17/Treg cell ratios are observed in some immunoin- flammatory diseases, e.g. sarcoidosis 8.
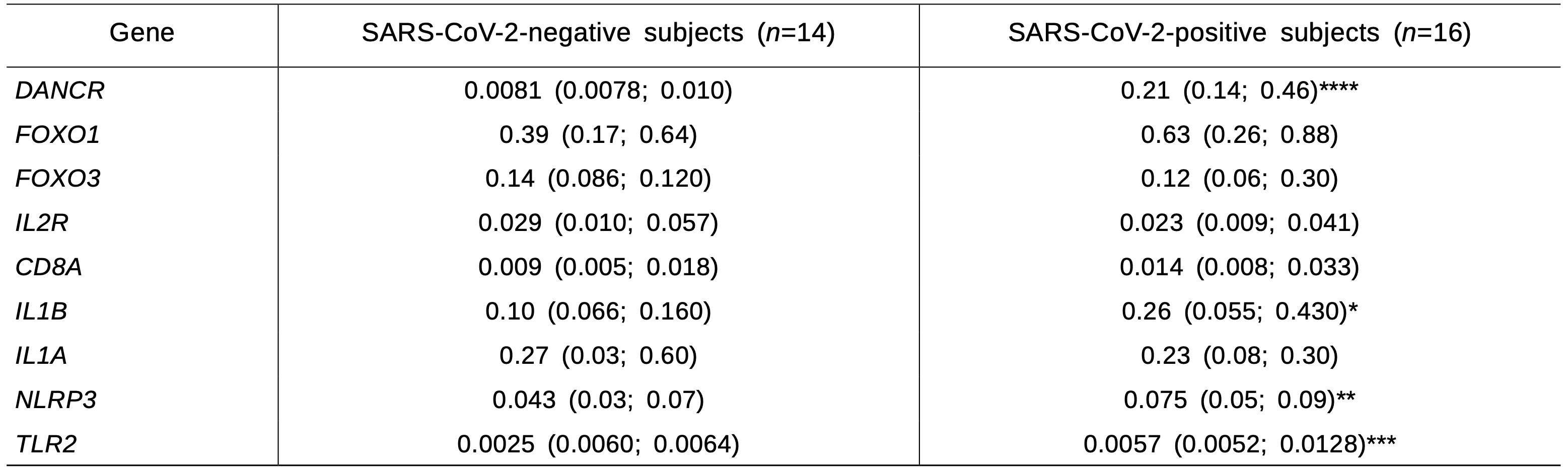
The mRNA level of long non-coding RNA (ln- cRNA) of DANCR, IL1B, NLRP3, and TLR2 genes in PBL of SARS-CoV-2-positive healthy donors was sig- nificantly higher than in subjects with negative status (Table 2). SARS-CoV-2 Nsp4 copy number correlated positively with the level of lncRNA DANCR transcripts (r=0.94; p<0.001) and negatively correlated with FOXP3 gene expression (r=-0.45; p=0.028). No differences in the content of transcripts of other studied genes were found.
The SARS-CoV-2 S-protein, in addition to the ACE2 protein, also binds to innate immune cell re- ceptors, such as Toll-like receptors (TLR) and NOD- like receptors (NLR) 9. This can lead to the initiation of signaling aimed at enhancing the expression of genes encoding the components of the NLRP3 inflam- masome and the formation of its active complex. It is likely that the presence of viral particles contributes to activation of TLR2 and NLRP3 inflammasome, as evidenced by an increase in the level of mRNA of the gene encoding these receptors, the NLRP3 gene (the cryopyrin gene, the main component of the NLRP3 inflammasome, which is involved in the production of mature form of the proinflammatory cytokine IL- 1β) and IL1B in SARS-CoV-2 PBL from conditionally healthy donors.
Fig. 1. Levels of FOXP3 (a) and RORγ (b) gene transcripts in PBL of healthy SARS-CoV-2-negative and SARS-CoV-2-positive subjects. *р=0.005, **р=0.0003.
lncRNA are involved in the regulation of inflam- matory reactions in response to a bacterial or viral infection. It was found that COVID-19 is also accom- panied by changes in the pattern of lncRNA expres- sion. In cells infected with SARS-CoV-2, an increase in the expression of 20 lncRNAs and a decrease in the level of 4 lncRNAs were observed 10. Enhanced expression of NEAT1 lncRNAs was also recorded in the lung tissues of patients with COVID-19 10. lncRNA DANCR is involved in the pathogenesis of inflammato- ry diseases 11. This lncRNA can be associated with regulation of FOXO transcription factors that, in turn, are involved in maintaining the transcriptional activity of the FOXP3 gene encoding the molecular marker of Treg cells. The decrease in FOXP3 gene expression in SARS-CoV-2-positive donors can be due to DANCR growth expression. In our study, no differences were found in the level of transcripts of the FOXO1 and FOXO3 genes in the compared groups. However, it is well known DANCR can affect activity and content of FOXO1 and FOXO3. Thus, lncRNA DANCR has been shown to reduce the FOXO1 content in macrophages by participating in the degradation of the ubiquitinat- ed form of this protein 12. DANCR can also inter- act with the adapter protein AU-binding factor 1 and increase the content of the FOXO3 protein without affecting its mRNA expression 13.
Thus, copies of SARS-CoV-2 viral particles can probably persist in PBL of patients for at least 5-6 months after recovery from COVID-19. The presence of SARS-CoV-2 Nsp4 transcripts in PBL of healthy individuals probably contributes to impair- ment of the suppressor properties of T-regulatory cells and enhancement of transcriptional activity of genes encoding proinflammatory proteins.
This work was performed within the framework of research project FMEN-2022-0017 12203100099-1 on scientific equipment of the Common Use Center of the Karelian Research Centre, Russian Academy of Sciences.
TABLE 2. Expression Level of Genes (relative units) Encoding Markers of T-Cell Differentiation and Proinflammatory Proteins in PBL of Healthy SARS-CoV-2-Negative and SARS-CoV-2-Positive Subjects
Note. *p=0.05, **p=0.02, ***p=0.003, ****p<0.0001.
REFERENCES:
1. Li S, Bai L, Dong J, Sun R, Lan K. Kaposi’s sarcoma-asso- ciated herpesvirus: epidemiology and molecular biology. Adv. Exp. Med. Biol. 2017;1018:91-127. doi: 10.1007/978- 981-10-5765-6_7
2. Bevova MR, Netesov SV, Aulchenko YS. The new coro- navirus COVID-19 infection. Mol. Gen. Microbiol. Virol. 2020;35(2):53-60. doi: 10.3103/S0891416820020044
3. Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA. 2021;118(21):e2105968118. doi: 10.1073/ pnas.2105968118
4. Easterbrook JD, Zink MC, Klein SL. Regulatory T cells enhance persistence of the zoonotic pathogen Seoul virus in its reservoir host. Proc. Natl Acad. Sci. USA. 2007;104(39):15 502-15 507. doi: 10.1073/pnas.0707453104
5. Jiang M, Guo Y, Luo Q, Huang Z, Zhao R, Liu S, Le A, Li J, Wan L. T-Cell Subset counts in peripheral blood can be used as discriminatory biomarkers for diagnosis and severity prediction of Coronavirus disease 2019. J. Infect. Dis. 2020;222(2):198-202. doi: 10.1093/infdis/jiaa252
6. Drago F, Ciccarese G, Rebora A, Parodi A. Human herpes- virus-6, -7, and Epstein-Barr virus reactivation in pityria- sis rosea during COVID-19. J. Med. Virol. 2021;93(4):1850- 1851. doi: 10.1002/jmv.26549
7. Pinto Lde F, Compri CM, Fornari JV, Bartchewsky W, Cintra DE, Trevisan M, Carvalho Pde O, Ribeiro ML, Velloso LA, Saad MJ, Pedrazzoli JJr, Gambero A. The im- munosuppressant drug, thalidomide, improves hepatic alterations induced by a high-fat diet in mice. Liver Int. 2010;30(4):603-610. doi: 10.1111/j.1478-3231.2009.02200.x
8. Huang H, Lu Z, Jiang C, Liu J, Wang Y, Xu Z. Imbal- ance between Th17 and regulatory T-Cells in sarcoidosis. Int. J. Mol. Sci. 2013;14(11):21 463-21 473. doi: 10.3390/ ijms141121463
9. Gadanec LK, McSweeney KR, Qaradakhi T, Ali B, Zulli A, Apostolopoulos V. Can SARS-CoV-2 virus use multiple re- ceptors to enter host cells? Int. J. Mol. Sci. 2021;22(3):992. doi: 10.3390/ijms22030992
10. Laha S, Saha C, Dutta S, Basu M, Chatterjee R, Ghosh S, Bhattacharyya NP. In silico analysis of altered expres- sion of long non-coding RNA in SARS-CoV-2 infected cells and their possible regulation by STAT1, STAT3 and interferon regulatory factors. Heliyon. 2021;7(3):e06395. doi: 10.1016/j.heliyon.2021.e06395
11. Zhang X, Ma L, Zhang C, Hou B, Zhou Y, Yu S. Silenc- ing LncRNA-DANCR attenuates inflammation and DSS- induced endothelial injury through miR-125b-5p. Gas- troenterol. Hepatol. 2021;44(9):644-653. doi: 10.1016/j. gastrohep.2020.10.008
12. Xie C, Guo Y, Lou S. LncRNA ANCR promotes invasion and migration of gastric cancer by regulating FoxO1 expression to inhibit macrophage M1 polarization. Dig. Dis. Sci. 2020;65(10):2863-2872. doi: 10.1007/s10620-019- 06019-1
13. Qian W, Cai X, Qian Q, Wang D, Zhang L. Angelica si- nensis polysaccharide suppresses epithelial-mesenchymal transition and pulmonary fibrosis via a DANCR/AUF-1/ FOXO3 regulatory axis. Aging Dis. 2020;11(1):17-30. doi: 10.14336/AD.2019.0512




.jpg)